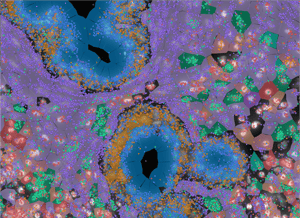
Spatial Transcriptomics
Spatial transcriptomics is the study of gene expression patterns within the context of tissue architecture and can provide a more complete understanding of cell-cell interactions. It is a powerful tool for anyone interested in understanding the complexity of biological systems and tissue organization at high resolution.
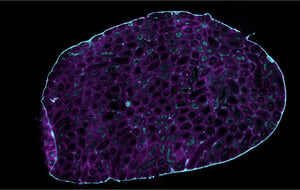
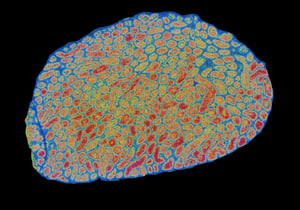
Our Spatial Multiomics Platforms

Off-the-shelf (up to 5K)
Custom Panels
Protein Sub-Panel Add-ons
Planning Your Spatial Experiment
There are several things to consider when deciding which spatial transcriptomics technology is best suited for your experiment. We put together this simple guide to help get you started.Single cell spatial imaging with 10x Genomics Xenium Analyser

We offer end-to-end Xenium service at competitive pricing, including:
- Sub-cellular resolution of individual transcripts
- Easy and intuitive tissue-scale comparisons
- Seamless panning, zooming, and analysis of your entire tissue section
- Off-the-shelf gene panels (100's to 1000's RNAs) and add-on Protein panels
Read More
Need Histology? We’ve Got You Covered.
The SAGC collaborates with the expert team at AMS Histology Services to offer a comprehensive spatial omics service from tissue to insight. Histology services at competitive rates for both internal and external clients.
Visium & Visium HD
Visium HD WT Panel (Human, Mouse, or Custom)
- Specialised slides contain spatially barcoded spots giving you high-resolution from standard tissue sectioning and staining.
- Compatible with FFPE and fresh frozen (FF) tissue samples.
- Whole transcriptome analysis – no need to select targets or specific region of interest.
- CytAssist enabled to allow transfer of tissues from archived slides.
Read More
NEW Visium HD 3’ Poly A (FF only)
- Species agnostic (enables plants and non-model organisms)
- Potential for TCR & BCR sequencing

Stereo-seq (SpaTial Enhanced Resolution Omics-Sequencing)
Whole transcriptomics for discovery applications (range of 1000-2000 genes/cell) leverages in situ mRNA capture and cDNA sequencing with barcoded spatial coordinates to create a spatial transcriptomic profile of the tissue section:
- Subcellular spatial localisation of mRNA
- Species agnostic
- Spot size = 200 nm diameter
- Range of field size to accommodate larger tissue samples (up to 13 cm x 13cm)
STOmics Stereo-seq OMNI for FFPE Samples
- Total RNA and Microbial RNA
- Enables Host Pathogen Detection
- Species Agnostic
- Single-cell Resolution
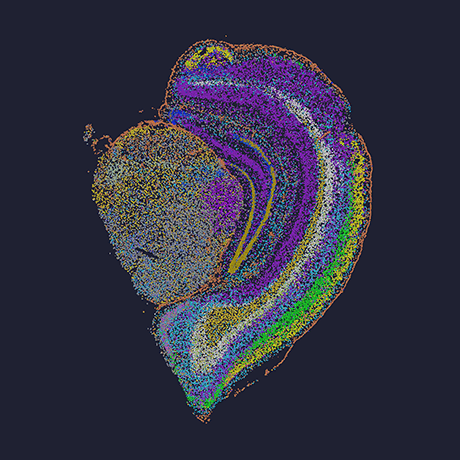
Xenium In Situ
Targeted gene expression and protein quantification at subcellular resolution
Xenium is an end-to-end platform that enables high-throughput subcellular mapping of RNA targets.
- Menu of off-the-shelf gene panels (up to 5K) and add your custom gene targets using free panel designer
- Compatible with fresh frozen (FF) and formalin-fixed, paraffin-embedded (FFPE) tissues
- Microscopy based read-out (no NGS), allows H&E post-run
- Single molecule RNA and Protein targets (subcellular resolution)

CosMx SMI
Imaging-based in situ analysis platform delivering spatial multiomic profiling from FFPE and FF samples to subcellular resolution.
- High-plex RNA panels: quantification and visualisation of 3 sizes of human RNA panels; 1K, 6K, 19K WTX (human whole transcriptome, mouse WTX available soon).
- Standalone protein or same-slide add-onto RNA assays for multiomic analyses.
- CosMx v2.0 upgrade delivers 1.5 to 2x sensitivity increases and best in class cell segmentation.

"A previously inaccessible view of tissue biology”.
Prof. David Lynn, SAGC Scientific Director



